S3140
Using AI to Characterise Unknown Bacteriophage from Poorly-Performing vs Well-Performing Broiler Flocks
Dr Didier Devaurs (University of Strathclyde), Dr Timofey Skortsov (Queen's University Belfast), Dr Leighton Pritchard (University of Strathclyde), Dr Victoria Smyth (Agri-Food and Biosciences Institute -AFBI).
Entry:
Cohort 3/October 2026
Interview Date:
Wednesday, 26th November (PM)
Eligibility:
Accepting Home & International Applications
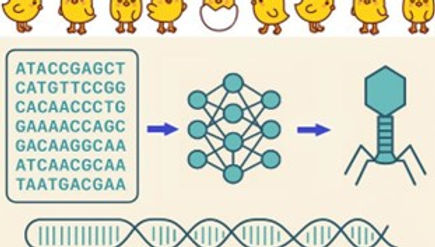
Although bacteriophage can combat many bacterial diseases, they constitute an under-utilised resource because most species remain unidentified. Bacteriophage offer great hope for future “One Health” treatment options, provided the groundwork is done to establish their identities via host infection. However, traditional experimental laboratory methods for bacteriophage identification are slow and labour-intensive. With predictions of health issues soon to rise sharply due to increasing antimicrobial resistance, both in humans and animals, novel high-throughout computational tools for bacteriophage identification are needed. This project will explore bacteriophage applications to improve animal health in the poultry industry, a critical commercial sector in the UK agri-food system.
As this project is fully interdisciplinary, the appointed student will apply a combination of computational methods andexperimental techniques. They will first use classical computational sequence-based tools from bioinformatics to identify bacteriophage DNA from previously collected poultry samples. Then, they will develop novel AI-based computational methods for bacteriophage identification (e.g., taxonomic labelling) and characterisation (e.g., functional gene identification). These computational tasks, carried out at the University of Strathclyde, will constitute most of the project. Then, the student will apply gold-standard experimental techniques at Queen’s University Belfast and AFBI, to isolate bacteriophage and screen their potential bacterial hosts/targets.
The appointed student will receive cross-disciplinary training in machine learning, software development, avian virology and molecular microbiology. They will start on the project by learning how to use and develop computational methods involving cutting-edge deep learning techniques, such as large nucleotide/protein sequence models. Later in the project, they will work closely with stakeholders in the field, from microbiologists to veterinarians in the UK and international poultry sector. They will emerge with cutting-edge skills and experiences in deep learning, computational biology, and microbiology, which are all in strong demand in academia, as well as in the public and private sectors.
